Metabolites, Free Full-Text
Por um escritor misterioso
Descrição
This research focused on establishing a hierarchy concerning the influence of various biological markers and body composition parameters on preventing, diagnosing and managing Metabolic Syndrome (MetS). Our cross-sectional cohort study included 104 subjects without any atherosclerotic antecedent pathology, organized in two groups (with and without MetS). All participants underwent clinical and anthropometric measurements, DEXA investigation and blood tests for all MetS criteria, together with adiponectin, leptin, insulin, uric acid and CRP. Based on mathematical logic, we calculated a normalized sensitivity score to compare the predictive power of biomarkers and parameters associated with MetS, upon the prevalence of MetS. Patients with MetS report higher levels of uric acid (p = 0.02), CRP (p = 0.012) and lower levels of adiponectin (p = 0.025) than patients without MetS. The top three biological markers with the highest predictive power of the prevalence of the disease are HDL, insulin, and adiponectin:leptin ratio, and the top three body composition parameters are trunk fat-free percentage, waist-height ratio and trunk fat percentage. Their high sensitivity scores differentiate them from all the other markers analysed in the study. Our findings report relevant scores for estimating the importance of cardiometabolic risks in the prevalence of MetS. The high rank of protective markers, HDL and trunk fat-free percentage, suggest that positive effects have a stronger association with the prevalence of MetS, than negative ones do. Therefore, this risk stratification study provides important support for prevention and management programs regarding MetS.

MetNet: Metabolite Network Prediction from High-Resolution Mass Spectrometry Data in R Aiding Metabolite Annotation

Metabolism and epigenetics at the heart of T cell function: Trends in Immunology

Multi-omics-based label-free metabolic flux inference reveals obesity-associated dysregulatory mechanisms in liver glucose metabolism - ScienceDirect
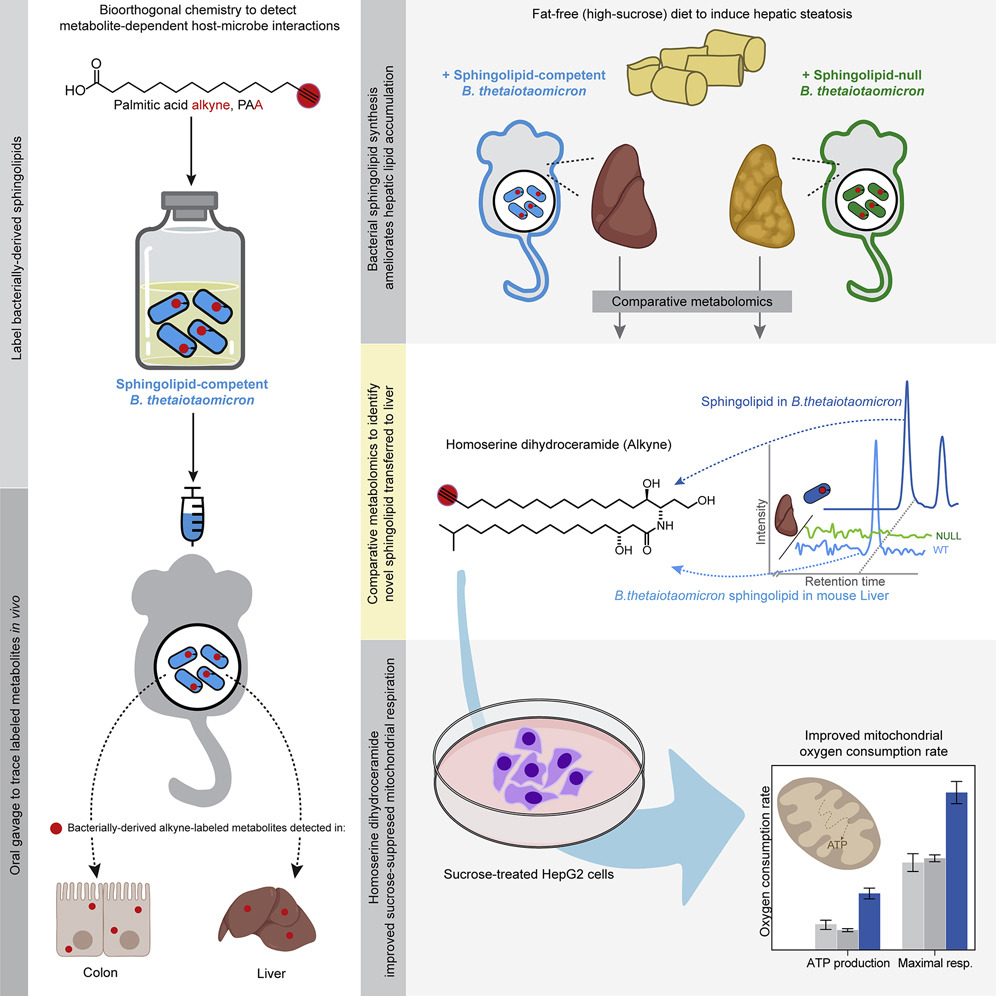
Cell Host & Microbe on X: Gut #microbiota sphingolipids modulate hepatic metabolism. @henrychembio @MinTingLee1 @lizljohnson ID & trace unique gut bacterial sphingolipid to colon & liver. Bacterial sphingolipid synthesis reduces hepatic lipid

The American Journal of Pathology
de
por adulto (o preço varia de acordo com o tamanho do grupo)